It's Not My Responsibility To Fix Your Broken Datasets And Pipelines...
I’ve been playing with some public datasets recently. In particular looking at datasets provided by various vendors. Sometimes these datasets are not super-well documented… or don’t seem to follow basic standards.
Here’s what I expect when I’m looking for a benchmark dataset for a DNA sequencing platform:
A set of fastq files.
If these are for the now popular Genome-in-a-bottle samples I expect one of them to have HG0002 in the filename.
That’s it really… if there’s anything else there’s going to be confusion. In Oxford Nanopore’s case1:
There are fastqs in the dataset, in a directory with HG002 in the path. But they are the wrong files to use… don’t use those. And you should just know that the only fastqs available are not ones you should use.
There are 2 different sets of cram files. So if I want fastqs I’ll need to figure out which reference to use and extract fastqs from these I guess?2
You can basecall the data yourself. But don’t choose the wrong model because that will be wrong too.
In fact one of the models (the fast model) should never be used by anyone ever.
Look, I get it, there may be limited use cases for a fast model that provides quick feedback but poor results. They are very limited use cases. Being able to run the fast model should probably be a compile time option or something that requires agreeing to a big obvious warning. Or setting an environment variable.
Don’t just present the user with 3 different models two of which have non-descriptive names (hac and sup) and expect them to choose the one you want them to choose.
For a blog post, I’m just going to pick whatever seems the most obvious choice (the fast model or the fastqs provided) stick it on the Internet and see if someone corrects me3.
If I was being paid to evaluate the platform I might be more systematic. A user casually evaluating your technology? Probably they’d just do what I did… in any case:
I don’t accept the responsibility to fix your broken datasets. Any more than anyone has a responsibility to read my posts.
ONT are not the only vendor to blame here. I’m now looking at Ultima data… before downloading their data I’m asked to agree to this nonsense:
So I guess I’ll be sending a postmarked letter to Ultima4 to opt-out of arbitration in order to download the data5.
In the repository itself I’m presented with multiple pdfs and it seems cram files… do I have to read the pdf to figure out which reference was used? And then figure out how to convert the crams to fastq using that reference6?
Oh cool there’s a link to the reference on the original page… weird that its not in the repository but whatever… oh…
What a pain… why not make it easy. At least that way I’ll start my analysis not already annoyed…
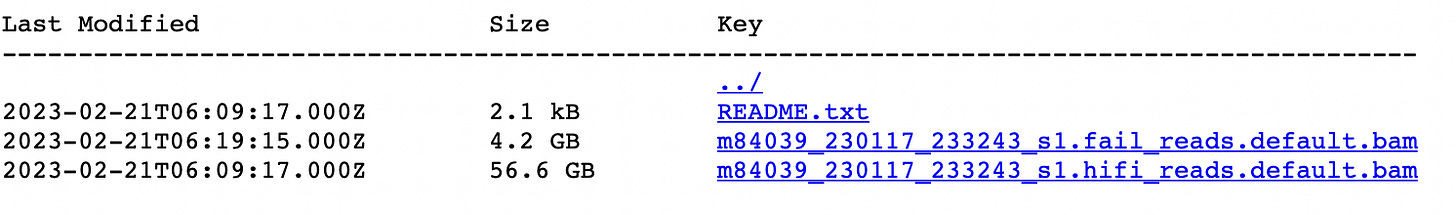
I’ll contrast both these examples with one positive example. I wanted a PacBio dataset. So I went to the Pangenome Consortium data page. Clicked the big “PacBio HG002 Data” button and was immediately taken to this page:
Two BAM files. One “hifi”, one “fail”. It’s pretty obvious which one I should use. And it’s reasonably easy to convert the BAMs to fastqs if I want to! I stick the BAM though the standard tools and a more or less sensible output pops out! Easy!
So…
While I’m just some random blogger, this is all going to apply to researchers and companies evaluating vendor products too. Making things easy for your users seems like it’s probably in your best interests…
TLDR;
I don’t view it as my responsibility to fix your broken datasets and pipelines.
At the time I did the analysis, they may have updated it now…
At the time of writing the page contains no information on which reference was used to generate the cram files. Perhaps samtools will try and download it /if/ it can match the MD5 in the cram file… (files don’t seem to contain an embedded reference). We can only hope.
Nope samtools view hangs on my system…
We can use samtools head to find out where the reference came from:
/scratch/cwright/grch38/GCA_000001405.15_GRCh38_no_alt_analysis_set.fasta
Cool! We just need access to cwright’s computers to get a copy.
It also references this file: GCA_000001405.15_GRCh38_no_alt_analysis_set.
fna
Probably if we randomly google for this file it’ll be fine… what a mess. Grab this:
https://www.encodeproject.org/files/GRCh38_no_alt_analysis_set_GCA_000001405.15/
Trying to process the cram with Dorado results in this failure (even if provided with a reference):
[W::find_file_url] Failed to open reference "https://www.ebi.ac.uk/ena/cram/md5/6aef897c3d6ff0c78aff06ac189178dd": Protocol not supported
[E::fai_build3_core] Failed to open the file /scratch/cwright/grch38/GCA_000001405.15_GRCh38_no_alt_analysis_set.fasta
[E::refs_load_fai] Failed to open reference file '/scratch/cwright/grch38/GCA_000001405.15_GRCh38_no_alt_analysis_set.fasta'
[E::cram_get_ref] Failed to populate reference for id 0
[E::cram_decode_slice] Unable to fetch reference #0:10001-134934So I’ll have to convert it to a BAM or fastq… this won’t work with a compressed reference… so I’ll also have to decompress the reference…
This is going to be even more true if you provide non-standard and biased metrics in your performance claims, like modal read accuracy (which nobody else uses). In this case, I won’t be interested in generating your biased metric. And the standard tools won’t give me this metric so I’ll have nothing to verify your metrics against.
You could just download it all directly without agreeing to those terms I guess?
aws s3 cp s3://ultima-ashg-2023-reference-set/crams/030945-NA24385-Z0114-CAACATACATCAGAT.cram . --no-sign-request
gsutil cp gs://gcp-public-data--broad-references/hg38/v0/Homo_sapiens_assembly38.fasta .
Someone seemed to suggest these contained an embedded reference. They do not appear to contain an embedded reference:
samtools reference -e 030945-NA24385-Z0114-CAACATACATCAGAT.cram
CRAM file has slice without embedded reference
If I attempt to extract the CRAMs without a reference as follows:
samtools fastq -1 out.R1.fastq -2 out.R2.fastq 030945-NA24385-Z0114-CAACATACATCAGAT.cram